Kanvas Bioscience wants to revolutionize microbial research and infectious disease diagnostics with next generation imaging technology that is able to provide deeper information on the spacial organisation and behaviour of microbial communities.
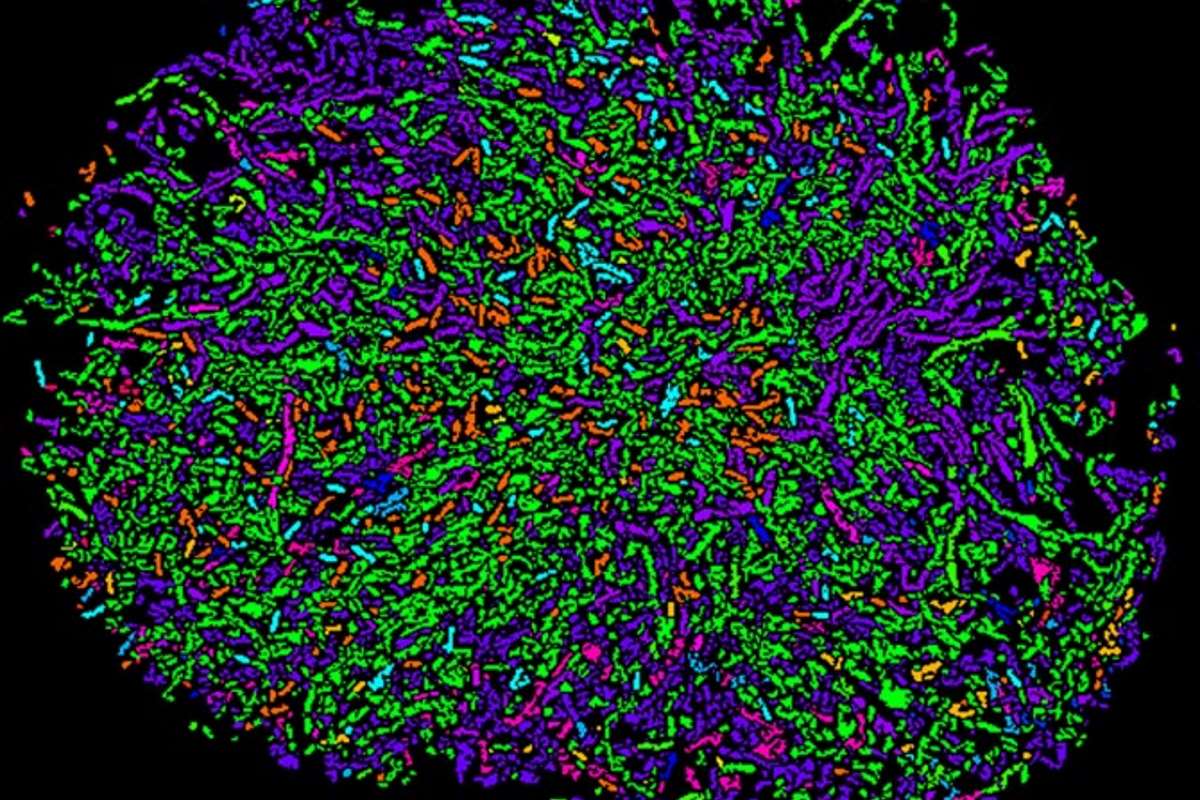
The company’s highly multiplexed imaging technology originates from Cornell University and uses a two-step process called high phylogenetic resolution microbiome mapping by fluorescence in situ hybridization (HiPR-FISH). HiPR-FISH provides a framework for analyzing the spatial organization of microbial communities in tissues and the environment at single cell resolution.
To locate the microbial communities, the researchers designed oligonucleotide probes that target specific bacteria cells based on the presence of a signature gene sequence and they made another group of probes that label the cells with fluorophores. Then the team used confocal microscopy to light up the fluorescent markers with lasers, and they used machine learning and custom software to decode the fluorescence spectra and interpret the images, resulting in what they describe as an efficient and cost-effective technology with single-cell resolution.
The technology has been applied to two different systems: the gut microbiome in mice and the human oral plaque microbiome. In the case of the gut microbiome, the team were able to demonstrate how the spatial associations between different bacteria are disrupted by antibiotic treatment.
Spatial mapping could potentially be an important tool for studying and possibly treating a range of diseases in which bacteria are a major culprit, such as inflammatory bowel disease, colorectal cancer and infection.
